Phytoplankton dynamics
Time series analysis
In collaboration with Ifremer Arcachon, we work on phytoplankton community dynamics, especially diatoms (one of the most abundant classes of plankton, actively involved in biogeochemical cycles).
We are particularly interested in:
- Drivers of blooms
- Coexistence mechanisms
Our research combines long-term data with statistical and theoretical models.
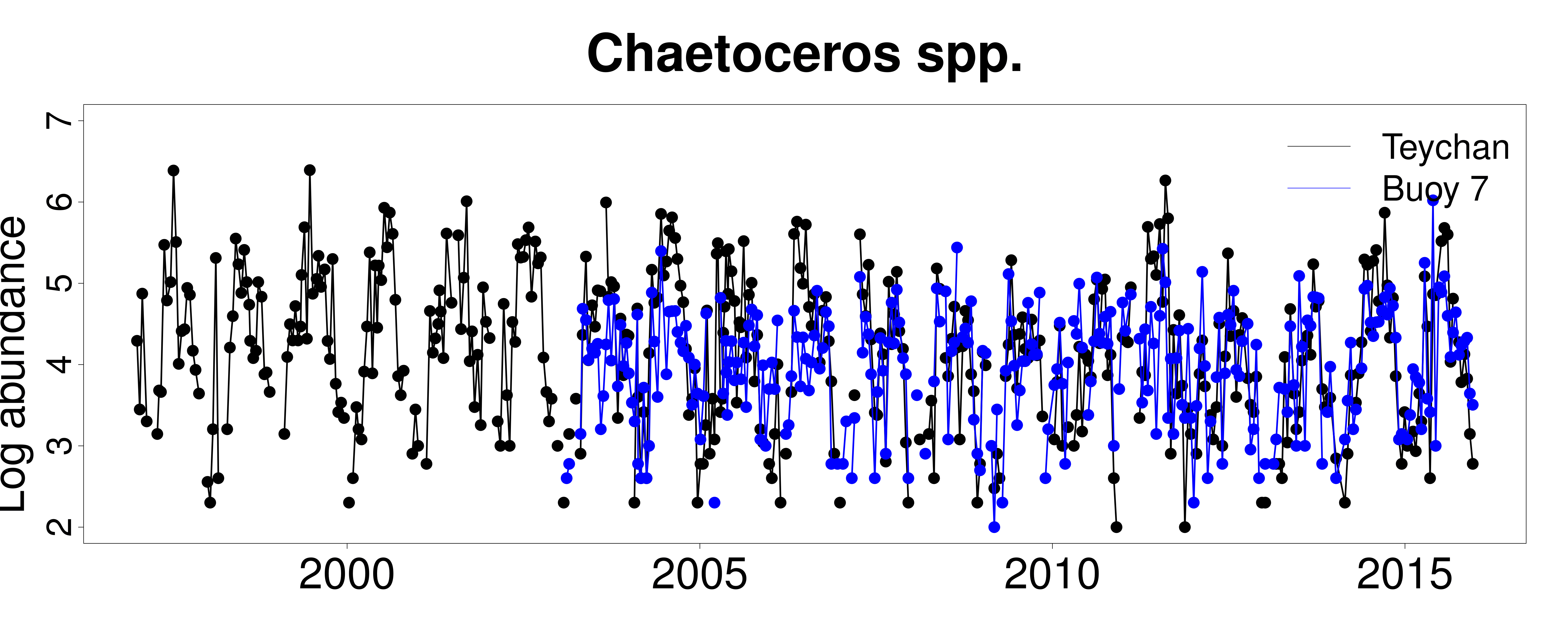
Our analyses of the time series in Arcachon Bay can be found in Barraquand et al. 2018, Oikos. This work has been extended to the French coastline in Picoche et al. 2019.
Metabarcoding
We are also involved in a metabarcoding phytoplankton inventory project funded by Labex COTE (MALABAR), with BioGeCo (INRA, University of Bordeaux), Ifremer and EPOC (CNRS, University of Bordeaux).

Usually phytoplankton counts and identification are made by optical microscopy.
The MALABAR project (Metabarcoding Applied to the Biodiversity of Microalgal Communities in the Arcachon Bay) aims to use new molecular-based approaches (metabarcoding) in addition to the usual inventories. We hope to have another view of the phytoplankton biodiversity, especially for smaller size classes which are not easily seen with classical methods.
A sampling campaign was carried out over a year. We sampled the water surface and sediments at 4 points, for each of the 4 seasons, at low and high tide.
These samples are currently being sequenced for two markers (8S-v4 and rbcL). The taxonomical identification of the sequences will be done by the search for similarity in two planktonic databases (Thonon diatom database, TARA database). However, we have already estimated through a first survey that about 60% of sequences can not be identified with this method. This is why we will also compute OTUs (Operational Taxonomic Units, i.e. clusters of genetically related individuals). This will allow us to assign a taxonomy to unknown sequences based on the genetic distance in the same cluster or between clusters.